A major aspect of plant cultivation involves the prevention, management, and treatment of diseases. This often occurs through a combination of means, such as specialised hygiene protocols, genetic selection, and biological and chemical control. Combined, these methods can be extremely successful at mitigating pests and diseases, however; some diseases are insensitive to chemical agents or may develop resistance over time. When this occurs, plant disease maintenance can become challenging and diseases have a greater chance of becoming established, often resulting in major yield losses for those plants that are affected.
This is an exciting development in understanding not only the molecular defence systems in hop but also the defences of closely related species (such as its closest relative Cannabis sativa) and plants more broadly!

Hop (Humulus lupulus) powdering mildew (Podosphaera macularis) is an example of an agricultural pest lacking effective means for control, and as such results in major annual losses to hop farmers globally. Whilst chemical control agents continue to be developed/ tested, other groups are exploring ways to boost the plants natural defence systems. This often focuses on identification of genes which are associated with increased disease resistance which can then be targeted for selective breading or genetic modification to improve crop performance (through disease resistance). These genes could be enzymes or antimicrobial agents to target pathogen-specific compounds, such as chitinases which break down chitin, a sugar complex present in fungi and insects (but absent from plants).
Nutrifield R&D has worked collaboratively with RMIT University to confirm the gene encoding the putative antimicrobial (chitinase) protein in hop, known as hop chitinase 1 (HCH1). These findings were published in the journal Physiological and Molecular Plant Pathology, and titled “Bioinformatic analysis of the putative hop antifungal chitinase HCH1”. This is an exciting development in understanding not only the molecular defence systems in hop but also the defences of closely related species (such as its closest relative Cannabis sativa) and plants more broadly!
The project utilised a branch of science known as bioinformatics; wherein in silico (computer based) tools such as software and online databases are used to analyse genetic and proteomic sequences. A range of bioinformatic tools were used to analyse the proposed HCH1 gene and compare it and it’s translated protein sequence, to other characterised chitinase genes and proteins in related species. This process identified conserved sequences, which corresponded to functional regions including a chitinase binding domain and a glycoside hydrolyse domain; more simply, a region which binds chitin and a region which breaks it.
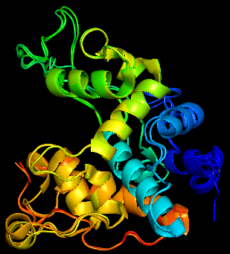
Finally, bioinformatic tools were used to develop predicted structural models for the protein and then compare this predicted structure to other characterised protein structures. This found that the predicted protein had a high degree of similarity with other plant-derived chitinase proteins. Combined, these results identified both structural and functional similarity to known chitinase proteins, confirming it’s (HCH1) identity as the hops chitinase.
The findings support the identity of the hops chitinase gene, and it’s targeting for selective breading for increasing hop disease resistance. It is also an encouraging step forward in identification of chitinase genes in other closely related species, such as its closest relative Cannabis sativa.
The complete results of this study were published in the journal Physiological and Molecular Plant Pathology, which can be found here: https://www.sciencedirect.com/science/article/abs/pii/S0885576520301831 – the title page has been copied in below.
Go check out our previous write up on our publication on the impact of biostimulants to root development in vegetative cuttings: https://www.nutrifield.com.au/research-development/comparing-biostimulants-to-optimise-root-formation-branching/